Submitted by Administrator on Tue, 30/07/2013 - 15:52
 A team of physicists working at the Universities of Cambridge and Leipzig have used a combination of single molecule techniques and theoretical calculations to follow and quantitatively describe the relaxation dynamics of DNA molecules. The work was published in the journal Nature Communications (http://www.nature.com/ncomms/journal/v4/n4/full/ncomms2790.html).
A team of physicists working at the Universities of Cambridge and Leipzig have used a combination of single molecule techniques and theoretical calculations to follow and quantitatively describe the relaxation dynamics of DNA molecules. The work was published in the journal Nature Communications (http://www.nature.com/ncomms/journal/v4/n4/full/ncomms2790.html).
DNA is the carrier of genetic information in every living organism, and its unique double-helical structure endows it with some fascinating physical properties. As a molecule, DNA resembles a very long thin wire: it can become as long as human hair while being around ten thousand times thinner. Such long DNA molecules are usually coiled up like spaghetti inside cells but can be stretched out to their full length in the laboratory.
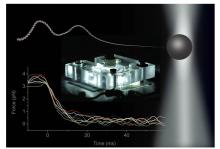
The group in Cambridge used laser optical tweezers to grab hold of one end of the DNA while the other end was stretched with a glass nanopore. This molecular tug of war can be used to accurately control the tension in the DNA molecule at the beginning of each experiment. After releasing one end of the DNA from the nanopore, the complex details of the relaxation dynamics of the DNA molecule cannot be directly observed in the microscope. But, using the theory developed in Leipzig, the process of snapping back can entirely be reconstructed from the measured force the other end of the DNA exerts onto the optical tweezers The precision of the experimental approach allows for a parameter-free comparison between measurements and theory.
The most notable characteristic of the DNA motion, as revealed by the measurements, is its surprisingly slow dynamics. During recoil, the DNA fragment, which has a length of about 10 microns, moves about 3 times slower than a rigid rod of the same length, and almost 20 times slower than it would in its fully coiled up native state. This remarkable dynamic friction enhancement could even substantially be increased in the future, by using longer DNA fragments. Its origin lies in the Brownian motion of the flexible thread, which is due to the random collisions with the surrounding water molecules. How a coherent deterministic motion of the DNA emerges from these incoherent thermal kicks is another interesting aspect of the theory tested in the experiment. Inspired by the widespread use and success of static single-molecule force spectroscopy techniques in recent years, the scientists hope that their novel time-resolved technique will in the future help to shed new light on the dynamics of DNA molecules and their interactions with proteins.
